57 Creating Spaghetti plots in GemPy
Contents
57 Creating Spaghetti plots in GemPy#
This notebook illustrates how to create spaghetti plots in GemPy illustrating the variety of model outcomes when performing probabilistic modeling in GemPy. This notebook was adapted from the work done here: https://github.com/elimh/stochastic-simulations-in-gempy.
Set File Paths and download Tutorial Data#
If you downloaded the latest GemGIS version from the Github repository, append the path so that the package can be imported successfully. Otherwise, it is recommended to install GemGIS via pip install gemgis and import GemGIS using import gemgis as gg. In addition, the file path to the folder where the data is being stored is set. The tutorial data is downloaded using Pooch (https://www.fatiando.org/pooch/latest/index.html) and stored in the specified folder. Use
pip install pooch if Pooch is not installed on your system yet.
[1]:
import warnings
warnings.filterwarnings("ignore")
import geopandas as gpd
import pandas as pd
import gempy as gp
import gemgis as gg
WARNING (theano.configdefaults): g++ not available, if using conda: `conda install m2w64-toolchain`
WARNING (theano.configdefaults): g++ not detected ! Theano will be unable to execute optimized C-implementations (for both CPU and GPU) and will default to Python implementations. Performance will be severely degraded. To remove this warning, set Theano flags cxx to an empty string.
WARNING (theano.configdefaults): install mkl with `conda install mkl-service`: No module named 'mkl'
WARNING (theano.tensor.blas): Using NumPy C-API based implementation for BLAS functions.
[2]:
file_path ='data/57_creating_spaghettii_plots_in_gempy/'
gg.download_gemgis_data.download_tutorial_data(filename="57_creating_spaghettii_plots_in_gempy.zip", dirpath=file_path)
Loading the Interface and Orientations Data#
The interface and orientation data is provided for a simple syncline model as Shapefile.
[3]:
interfaces = gpd.read_file('data/57_creating_spaghettii_plots_in_gempy/SynclineInterfaces.shp')
orientations = gpd.read_file('data/57_creating_spaghettii_plots_in_gempy/SynclineInterfaces.shp')
Deleting columns and dropping geometries#
[4]:
del interfaces['polarity']
del interfaces['dip']
del interfaces['azimuth']
interfaces['Z'] = -100
orientations['Z'] = -100
interfaces = pd.DataFrame(interfaces.drop(columns='geometry'))
orientations = pd.DataFrame(orientations.drop(columns='geometry'))
Inspecting DataFrames#
[5]:
interfaces.head()
[5]:
| X | Y | Z | formation | |
|---|---|---|---|---|
| 0 | 200 | 100 | -100 | Layer3 |
| 1 | 400 | 100 | -100 | Layer3 |
| 2 | 600 | 100 | -100 | Layer3 |
| 3 | 800 | 100 | -100 | Layer3 |
| 4 | 200 | 200 | -100 | Layer2 |
[6]:
orientations.head()
[6]:
| X | Y | Z | formation | polarity | azimuth | dip | |
|---|---|---|---|---|---|---|---|
| 0 | 200 | 100 | -100 | Layer3 | 1 | 0 | 45 |
| 1 | 400 | 100 | -100 | Layer3 | 1 | 0 | 45 |
| 2 | 600 | 100 | -100 | Layer3 | 1 | 0 | 45 |
| 3 | 800 | 100 | -100 | Layer3 | 1 | 0 | 45 |
| 4 | 200 | 200 | -100 | Layer2 | 1 | 0 | 45 |
Creating GemPy Model#
Initiate Model#
A relatively low resolution is chosen to speed up the modeling process.
[7]:
geo_model = gp.create_model('Model1')
[8]:
gp.init_data(geo_model, [0, 1000, 0, 1000, -500,0], [30,30,30],
surface_points_df = interfaces,
orientations_df = orientations,
default_values=True)
geo_model.surfaces
Active grids: ['regular']
[8]:
| surface | series | order_surfaces | color | id | |
|---|---|---|---|---|---|
| 0 | Layer3 | Default series | 1 | #015482 | 1 |
| 1 | Layer2 | Default series | 2 | #9f0052 | 2 |
| 2 | Layer1 | Default series | 3 | #ffbe00 | 3 |
Map Stack to Surfaces#
[9]:
gp.map_stack_to_surfaces(geo_model,
{"Strat_Series": ('Layer1', 'Layer2', 'Layer3')},
remove_unused_series=True)
geo_model.add_surfaces('basement')
[9]:
| surface | series | order_surfaces | color | id | |
|---|---|---|---|---|---|
| 0 | Layer3 | Strat_Series | 1 | #015482 | 1 |
| 1 | Layer2 | Strat_Series | 2 | #9f0052 | 2 |
| 2 | Layer1 | Strat_Series | 3 | #ffbe00 | 3 |
| 3 | basement | Strat_Series | 4 | #728f02 | 4 |
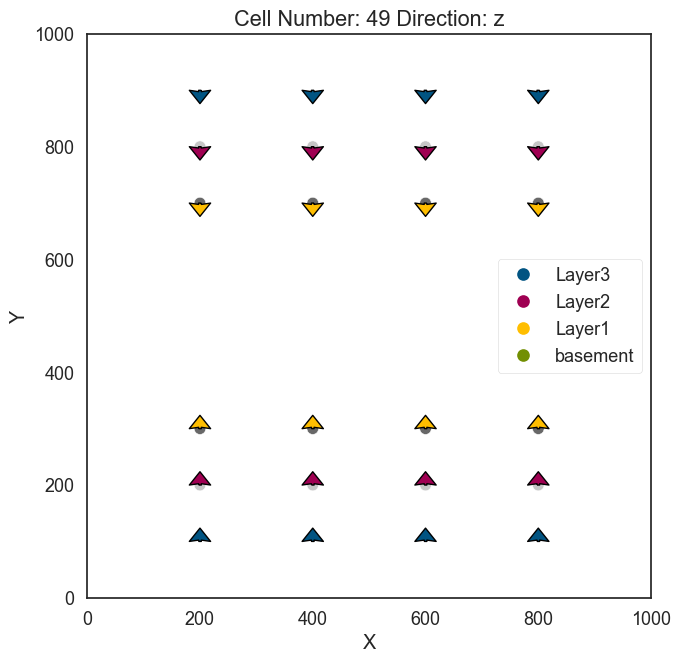
Plot Input Data#
[10]:
gp.plot_2d(geo_model, direction='z', cell_number = 49)
[10]:
<gempy.plot.visualization_2d.Plot2D at 0x1fd2af02560>
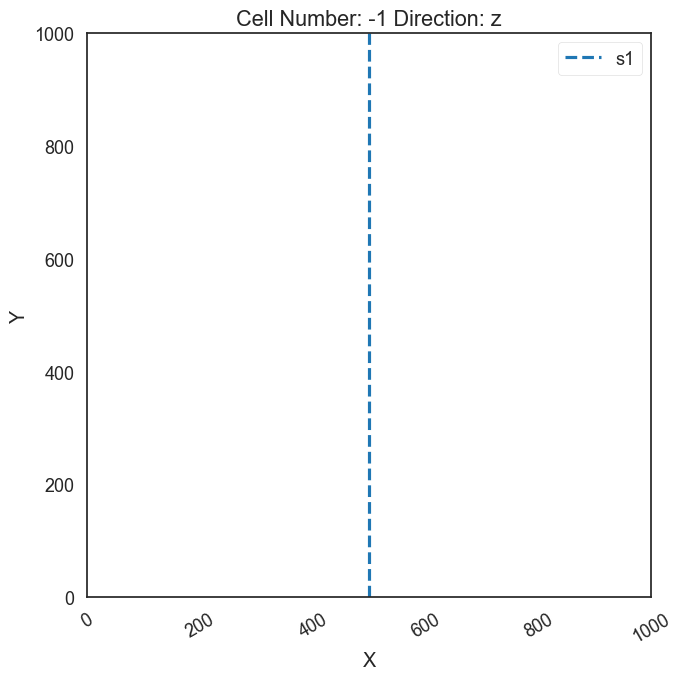
Creat custom section for Spaghetti Plot#
A custom section needs to be created to later obtain the Spaghetti Plot
[11]:
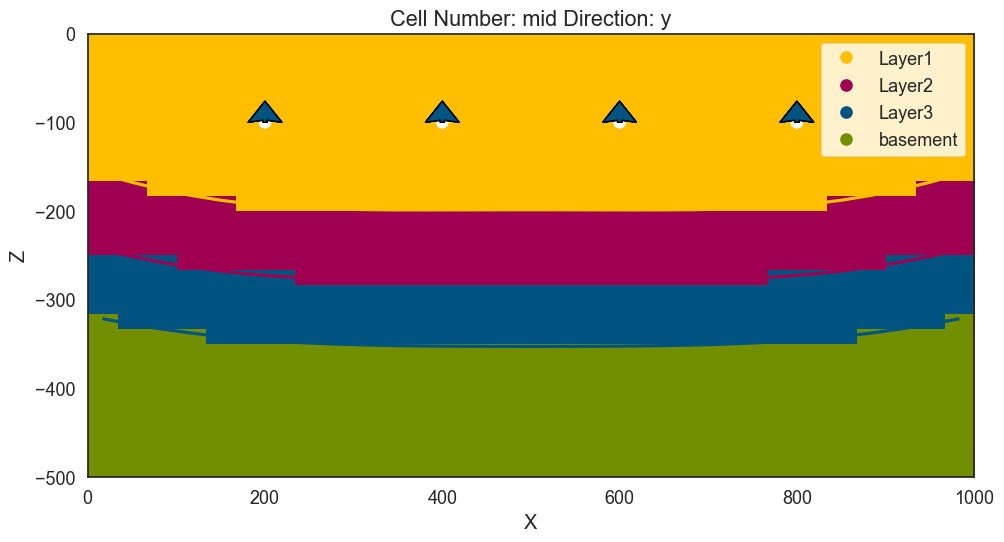
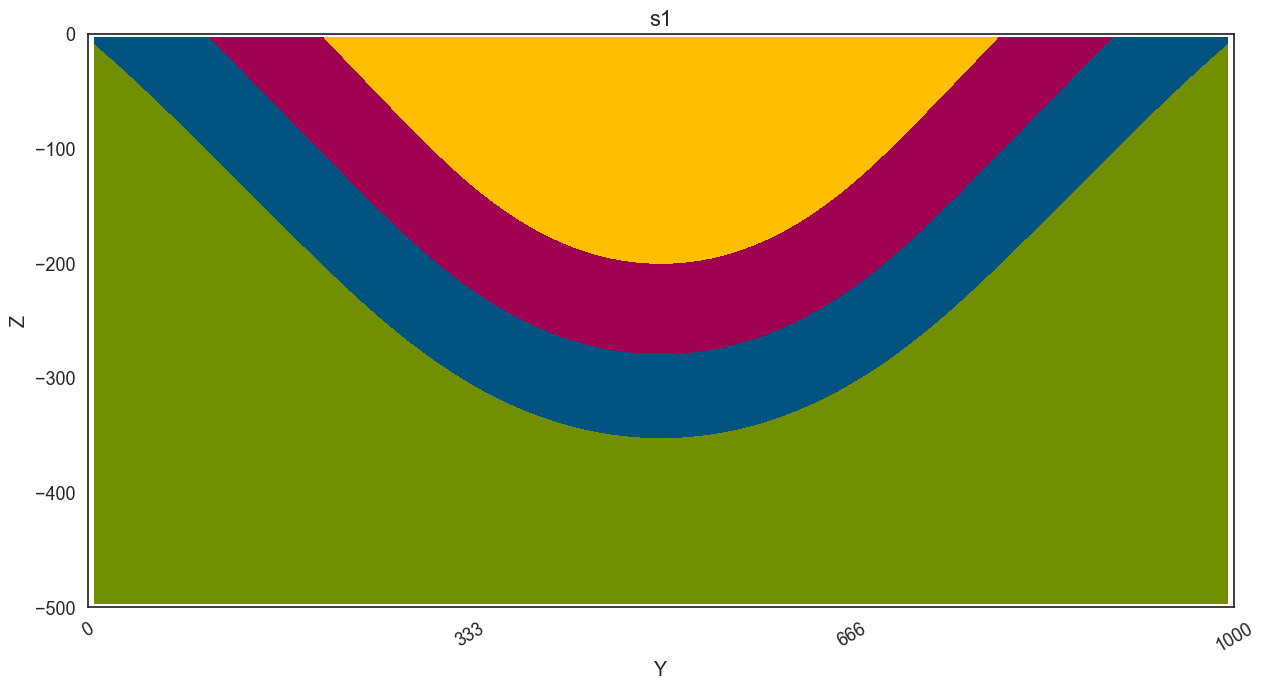
geo_model.set_section_grid({'s1':([500,0],[500,1000],[80,80])})
gp.plot.plot_section_traces(geo_model)
Active grids: ['regular' 'sections']
[11]:
<gempy.plot.visualization_2d.Plot2D at 0x1fd1f5dee60>
Set Interpolator#
[12]:
gp.set_interpolator(geo_model,
compile_theano=True,
theano_optimizer='fast_compile',
verbose=[],
update_kriging = False
)
Compiling theano function...
Level of Optimization: fast_compile
Device: cpu
Precision: float64
Number of faults: 0
Compilation Done!
Kriging values:
values
range 1500.00
$C_o$ 53571.43
drift equations [3]
[12]:
<gempy.core.interpolator.InterpolatorModel at 0x1fd293c2b60>
Compute Model#
[13]:
sol = gp.compute_model(geo_model)
Plot Model#
[14]:
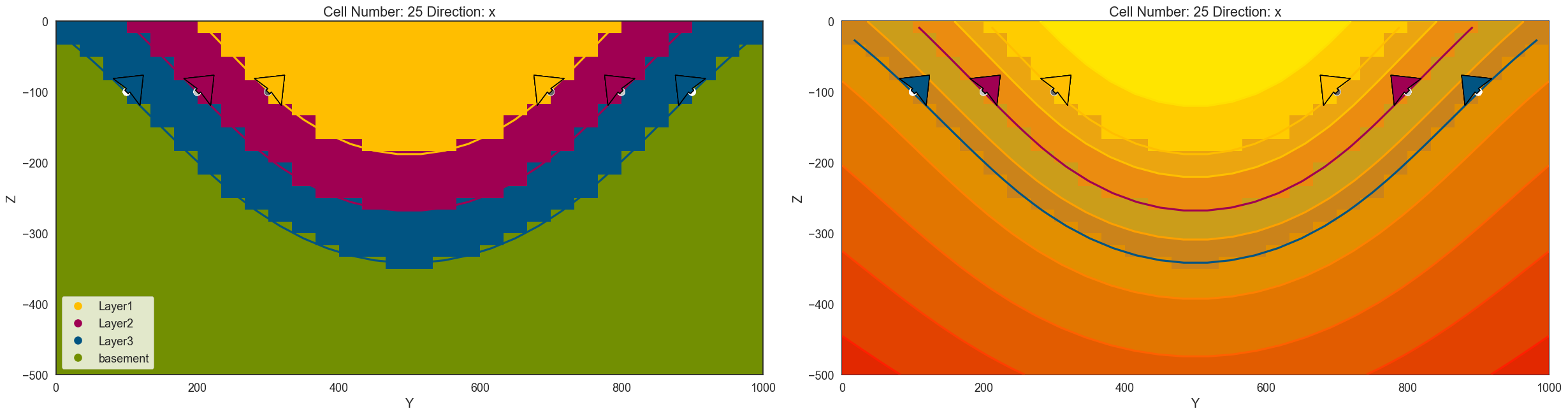
gp.plot_2d(geo_model, direction=['x', 'x'], cell_number=[25, 25,], show_data=True, show_scalar = [False, True])
[14]:
<gempy.plot.visualization_2d.Plot2D at 0x1fd2af01c90>
[15]:
gp.plot_2d(geo_model, section=['s1'])
[15]:
<gempy.plot.visualization_2d.Plot2D at 0x1fd2c5f11e0>
[16]:
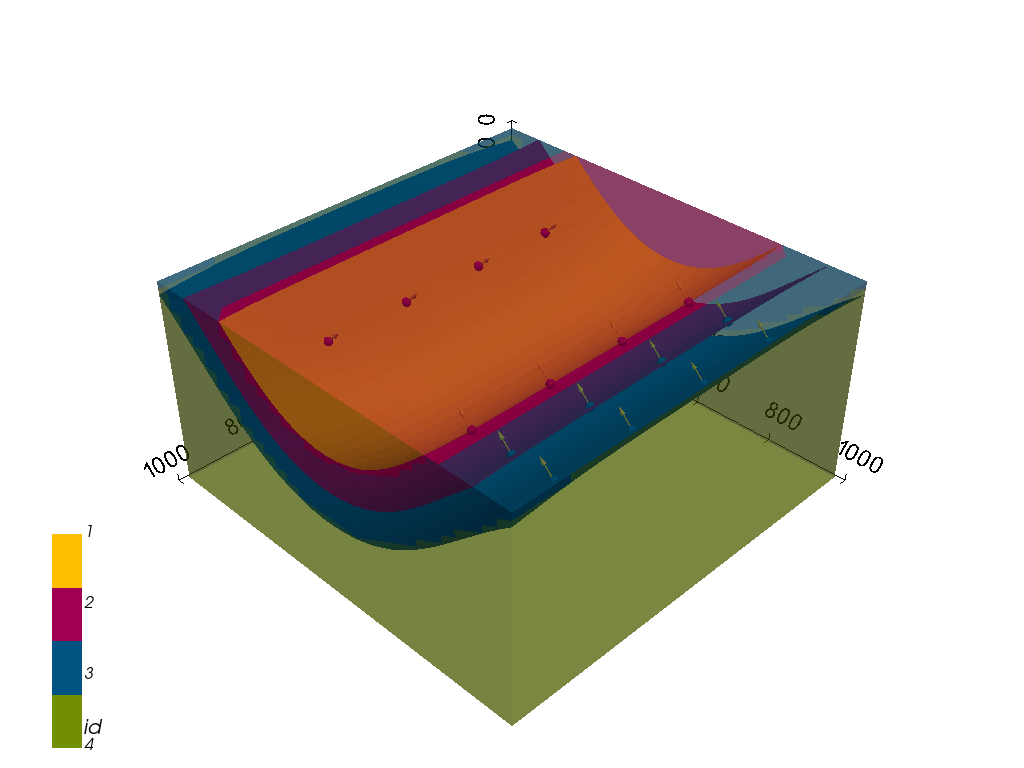
gpv = gp.plot_3d(geo_model,
image=False,
show_topography=True,
plotter_type='basic',
notebook=True)
Probabilistic Modeling#
During the probabilistic modeling, multiple model realizations will be created by varying the input parameter and recomputing the models with the new input data. The Monte Carlo approach is the base for the Spaghetti plot.
[17]:
from gempy.bayesian.fields import compute_prob, calculate_ie_masked
from gempy.core.grid_modules import section_utils
from tqdm.notebook import tqdm
import numpy as np
import copy
Getting polygondict for the spaghetti plot#
[18]:
polygondict_s1, cdict, extent = section_utils.get_polygon_dictionary(geo_model, 's1');
Copying the original model data#
A deep copy is created for all interface points and locations as well as their indices.
[19]:
df_int_X = copy.copy(geo_model.surface_points.df['X'])
df_int_Y = copy.copy(geo_model.surface_points.df['Y'])
df_int_Z = copy.copy(geo_model.surface_points.df['Z'])
df_or_X = copy.copy(geo_model.orientations.df['X'])
df_or_Y = copy.copy(geo_model.orientations.df['Y'])
df_or_Z = copy.copy(geo_model.orientations.df['Z'])
df_or_dip = copy.copy(geo_model.orientations.df['dip'])
df_or_azimuth = copy.copy(geo_model.orientations.df['azimuth'])
surfindexes = list(geo_model.surface_points.df.index)
orindexes = list(geo_model.orientations.df.index)
Monte Carlo Simulation#
[20]:
# Preventing matplotlib from plotting
%matplotlib agg
# Defining number of iterations
n_iterations = 50
# Getting indices
indices = list(geo_model.surface_points.df.index.sort_values()[:24])
indices_or = list(geo_model.orientations.df.index)
# Defining empty lith_block array
lith_blocks = np.array([])
# Defining empty list to store meshes
meshes = []
for iterations in tqdm(range(n_iterations)):
# Setting seed for reproducability
np.random.seed(iterations)
# Resetting Dataframes
geo_model.modify_surface_points(surfindexes, X=df_int_X, Y=df_int_Y, Z=df_int_Z)
geo_model.modify_orientations(orindexes, X=df_or_X, Y=df_or_Y, Z=df_or_Z,dip = df_or_dip, azimuth = df_or_azimuth)
geo_model.update_to_interpolator()
# Modifying Points
dip_sample = np.random.uniform(25,65, size=1)
for i in range(len(indices_or)):
geo_model.modify_orientations(indices_or[i], dip = dip_sample)
# Setting Interpolator and compute model
geo_model.update_to_interpolator()
sol = gp.compute_model(geo_model, compute_mesh=True)
#Spaghetti Plots
polygondict, cdict, extent = section_utils.get_polygon_dictionary(geo_model, 's1')
for form in polygondict.keys():
polygondict_s1.get(form).append(polygondict.get(form))
# Saving lith_block after each model run
lith_blocks = np.append(lith_blocks, geo_model.solutions.lith_block)
# np.save('lith_blocks_syncline_dip_all_new.npy', lith_blocks)
# Exporting meshes using GemGIS
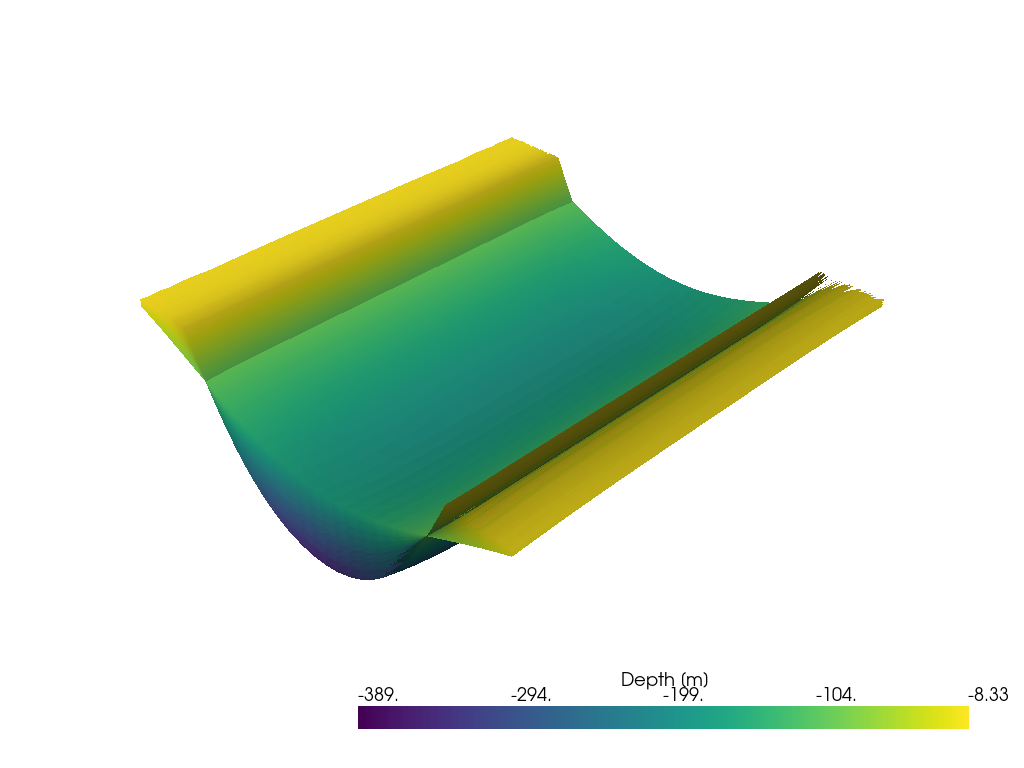
mesh = gg.visualization.create_depth_maps_from_gempy(geo_model, surfaces=['Layer2'])
meshes.append(mesh)
Reshaping lith_blocks array#
[21]:
# lith_blocks_syncline_dip_all = np.load('lith_blocks_syncline_dip_all.npy').reshape(n_iterations, -1)
prob_block_syncline_dip_all = compute_prob(lith_blocks.reshape(n_iterations, -1))
print(prob_block_syncline_dip_all.shape)
prob_block_syncline_dip_all
(4, 27000)
[21]:
array([[0. , 0. , 0. , ..., 0. , 0. , 0. ],
[0. , 0. , 0. , ..., 0. , 0. , 0.12],
[0. , 0. , 0. , ..., 0.5 , 0.64, 0.64],
[1. , 1. , 1. , ..., 0.5 , 0.36, 0.24]])
Plotting Probabilities#
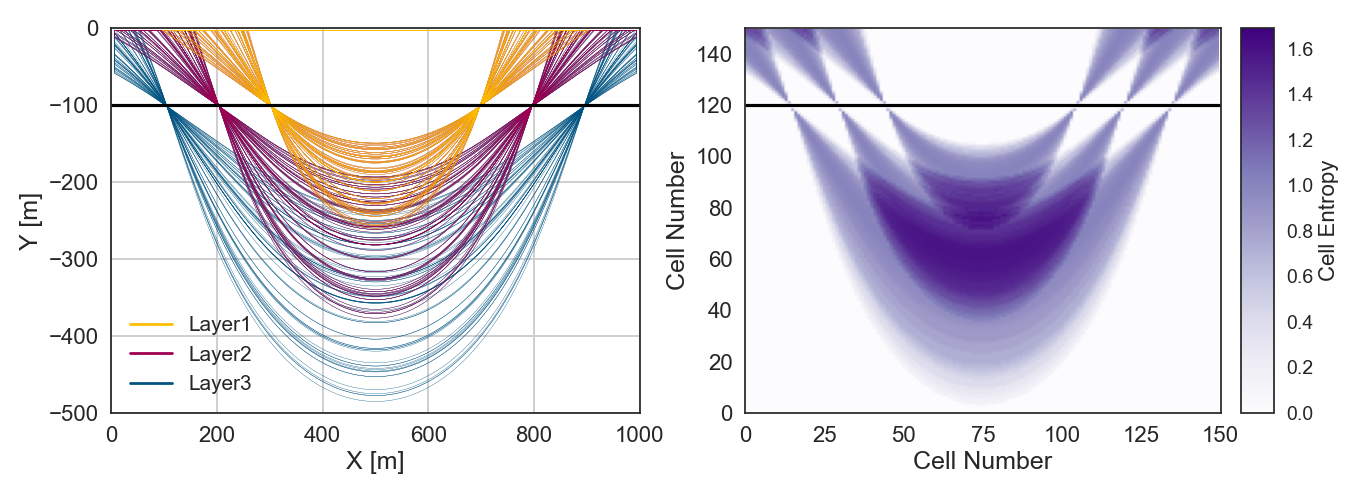
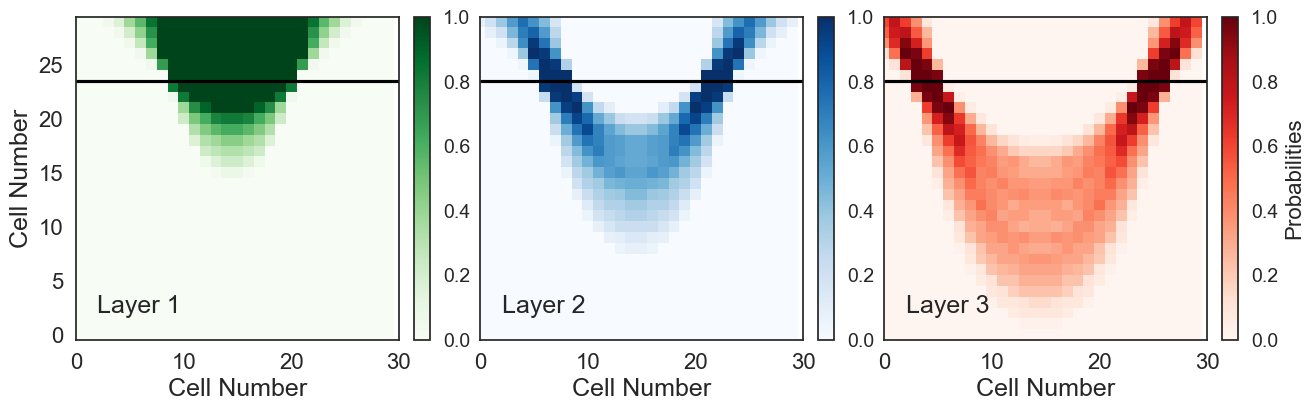
The probabilities of each layer being present in a cell are plotted.
[22]:
from mpl_toolkits.axes_grid1 import ImageGrid
import matplotlib.pyplot as plt
[23]:
def plot_probs(array):
fig = plt.figure(figsize=(15, 5))
grid = ImageGrid(fig, 111,
nrows_ncols=(1,3),
axes_pad=0.5,
share_all=True,
cbar_location="right",
cbar_mode="each",
cbar_size="5%",
cbar_pad=0.15,
)
cmaps = ['Greens', 'Blues', 'Reds']
for counter, ax in enumerate(grid):
im = ax.imshow(np.fliplr(array[counter].reshape(30,30,30)[
25].T), cmap=cmaps[counter], origin='lower', vmin=0, vmax=1)
ax.set_xlabel('Cell Number',fontsize=18 )
ax.set_ylabel('Cell Number',fontsize=18)
ax.hlines(y=23.5,xmin=0,xmax=200, color='k')
ax.set_xlim(0,30)
ax.text(x=2, y=2, s='Layer ' + str(counter+1))
cbar = ax.cax.colorbar(im)
ax.tick_params(axis='both', which='major', labelsize=16)
ax.tick_params(axis='both', which='minor', labelsize=16)
cbar.ax.tick_params(axis='both', which='major', labelsize=14)
cbar.ax.tick_params(axis='both', which='minor', labelsize=14)
ax.cax.toggle_label(True)
cbar.ax.set_ylabel('Probabilities', fontsize=16)
[24]:
%matplotlib inline
plot_probs(prob_block_syncline_dip_all)
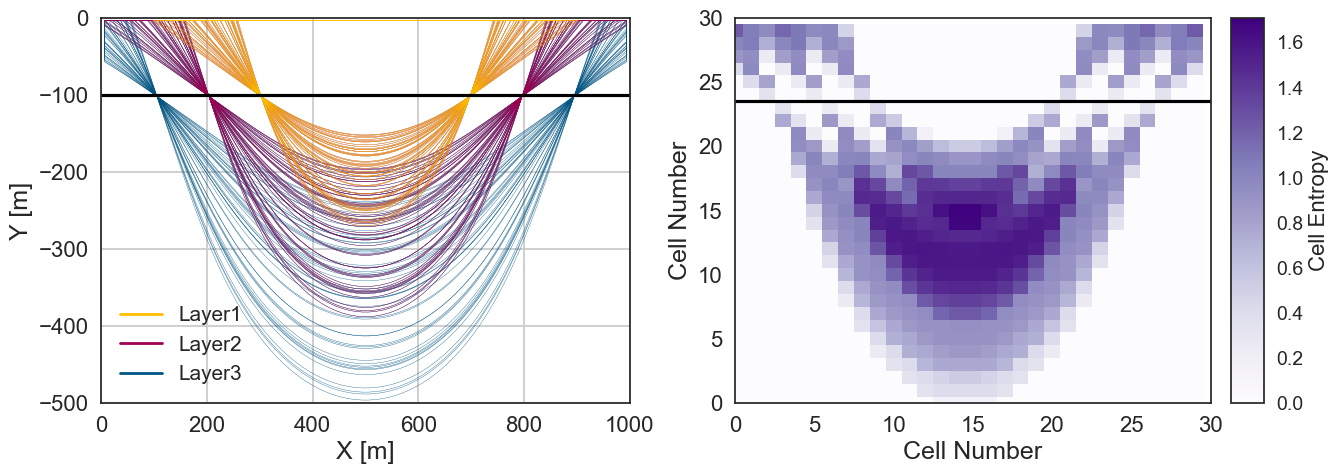
Plotting Entropy and Spaghetti Plot#
[25]:
import matplotlib.patches as patches
from matplotlib.lines import Line2D
from mpl_toolkits.axes_grid1 import make_axes_locatable
from matplotlib.path import Path
[26]:
%matplotlib agg
fig, ax = plt.subplots()
def plot_pathdict(pathdict, cdict, extent, ax=None, surfaces=None, color=None):
for formation in surfaces:
print(formation)
for path in pathdict.get(formation):
if path !=[]:
if type(path) == Path:
patch = patches.PathPatch(path, fill=False, lw=0.25, edgecolor=color)
ax.add_patch(patch)
elif type(path) == list:
for subpath in path:
assert type(subpath == Path)
patch = patches.PathPatch(subpath, fill=False, lw=0.25, edgecolor=color)
ax.add_patch(patch)
ax.set_ylim(-500,0)
ax.set_xlim(extent[:2])
[27]:
entropy_block_syncline_dip_all = calculate_ie_masked(prob_block_syncline_dip_all)
[28]:
%matplotlib inline
fix, ax = plt.subplots(nrows=1, ncols=2,sharex=False, sharey=False, squeeze=True , figsize = (15,5))
custom_lines = [Line2D([0], [0], color='#ffbe00', lw=2),
Line2D([0], [0], color='#9f0052', lw=2),
Line2D([0], [0], color='#015482', lw=2)]
plot_pathdict(polygondict_s1, cdict, extent, ax=ax[0], surfaces=['Layer3'], color=cdict['Layer3'])
plot_pathdict(polygondict_s1, cdict, extent, ax=ax[0], surfaces=['Layer2'], color=cdict['Layer2'])
plot_pathdict(polygondict_s1, cdict, extent, ax=ax[0], surfaces=['Layer1'], color=cdict['Layer1'])
ax[0].set_aspect('auto')
ax[0].hlines(y=-100,xmin=0,xmax=1000, color='k')
ax[1].set_xlim(0,1000)
ax[0].tick_params(axis='both', which='major', labelsize=16)
ax[0].tick_params(axis='both', which='minor', labelsize=16)
ax[0].set_xlabel('X [m]',fontsize=18)
ax[0].set_ylabel('Y [m]',fontsize=18)
ax[0].grid()
ax[0].legend(custom_lines, ['Layer1', 'Layer2', 'Layer3'], fontsize=15)
im = ax[1].imshow(np.fliplr(entropy_block_syncline_dip_all.reshape(30,30,30)[
15].T), cmap='Purples', origin='lower', vmax = entropy_block_syncline_dip_all.max(),aspect="auto")
ax[1].set_xlabel('Cell Number',fontsize=18)
ax[1].set_ylabel('Cell Number',fontsize=18)
ax[1].hlines(y=23.5,xmin=0,xmax=200, color='k')
ax[1].set_xlim(0,30)
ax[1].set_ylim(0,30)
ax[1].tick_params(axis='both', which='major', labelsize=16)
ax[1].tick_params(axis='both', which='minor', labelsize=16)
divider = make_axes_locatable(ax[1])
cax = divider.append_axes('right', size="7%", pad=0.2,)
cbar = fig.colorbar(im, cax=cax)
cbar.ax.tick_params(axis='both', which='major', labelsize=14)
cbar.ax.tick_params(axis='both', which='minor', labelsize=14)
cbar.ax.set_ylabel('Cell Entropy', fontsize=16)
fig.tight_layout()
plt.savefig('fig4.pdf', bbox_inches='tight', pad_inches=0.2)
Layer3
Layer2
Layer1
Plotting Meshes in 3D#
[29]:
meshes_layer2 = [mesh['Layer2'][0] for mesh in meshes]
meshes_layer2[0]
[29]:
| Header | Data Arrays | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
[30]:
import pyvista as pv
p = pv.Plotter(notebook=True)
for mesh in meshes_layer2:
p.add_mesh(mesh)
p.show()